Example of the autoblock usage#
Computational graphs in bosk requires their nodes to be a BaseBlock instances. To make user models including easier, the auto_block decorator was developed. Using it, you can easily convert your custom model to the BaseBlock. To get examples of converting scikit-learn interface models, see the bosk.block.zoo.models.classification subpackage code.
This exmaple requires the Pytorch to be installed. Make sure you have the torch package installed or run the following cell:
[ ]:
!pip install torch
[1]:
from bosk.block.auto import auto_block
from bosk.block.meta import BlockExecutionProperties
from bosk.executor.parallel.greedy import GreedyParallelExecutor
from bosk.painter.graphviz import GraphvizPainter
from bosk.pipeline.builder.functional import FunctionalPipelineBuilder
from bosk.stages import Stage
import torch
from torch.utils.data import TensorDataset, DataLoader
import numpy as np
from sklearn.datasets import make_moons
from sklearn.model_selection import train_test_split
from IPython.display import Image
Consider we have the following model:
[2]:
@auto_block(
execution_props=BlockExecutionProperties(cpu=False, gpu=True, plain=False, threadsafe=True),
random_state_field=None,
)
class NeuralNetwork(torch.nn.Module):
def __init__(self, dim: int) -> None:
super().__init__()
self.device = torch.device('cuda')
self.nn = torch.nn.Sequential(
torch.nn.Linear(dim, dim * 10),
torch.nn.ReLU(),
torch.nn.Linear(dim * 10, dim),
torch.nn.Tanh(),
torch.nn.Linear(dim, 1),
torch.nn.Sigmoid()
).to(self.device)
self.optimizer = torch.optim.Adam(self.nn.parameters(), 0.0005)
self.batch_size = 64
self.epochs_num = 500
self.loss_fn = torch.nn.CrossEntropyLoss()
def _get_tensor(self, x: np.ndarray) -> torch.Tensor:
return torch.from_numpy(x.astype(np.float32)).to(self.device)
def forward(self, x: torch.Tensor) -> torch.Tensor:
proba = self.nn(x)
return torch.concat((1 - proba, proba), dim=-1)
def transform(self, x: np.ndarray) -> np.ndarray:
with torch.no_grad():
x = self._get_tensor(x)
return self(x).cpu().numpy()
def fit(self, x: np.ndarray, y: np.ndarray) -> 'NeuralNetwork':
x_tens = self._get_tensor(x)
y_tens = self._get_tensor(y).long()
dataset = TensorDataset(x_tens, y_tens)
data_loader = DataLoader(dataset, self.batch_size, True)
self.train()
for _ in range(self.epochs_num):
for x, y in data_loader:
self.optimizer.zero_grad()
pred = self(x)
loss = self.loss_fn(pred, y)
loss.backward()
self.optimizer.step()
return self
Let’s describe what is happening here. We’ve got a Pytorch neural network model which we want to include in some bosk pipeline and train it on a GPU. To turn our model into a bosk BaseBlock we need to use auto_block decorator.
The decorator works with the classes having fit and transform methods. It uses the methods’ arguments names to create slots metainformation. Each argument name creates its own slot. If an argument is presented in the fit method, than the slot will be marked as used on the FIT stage. The same with the transform method and the TRANSFORM stage.
We can use auto_block without any arguments, and it will create a default CPU block with the default behaviour, which you can learn from the documentation. In our case we want this block to be marked to be runnable on the GPU (cpu=False, gpu=True), able to be parallelized (thread_safe=True), and as we didn’t implement neural network weights initialization according to the given random seed, we marked random_state_field=None to do nothing in the block.set_random_state method.
Now let’s make a pipeline with our model. Notice that we need to register our custom block in the functional builder. Also, it is needed to say that currently in bosk GPU data is stored using jax arrays, which are incompatible with the Pytorch tensors. This is why we didn’t put any MoveToBlock in the pipeline and implemented all the data transfer logic inside of the NeuralNetwork model.
[3]:
dim = 2
b = FunctionalPipelineBuilder()
X, y = b.Input()(), b.TargetInput()()
rf = b.RFC()(X=X, y=y)
rf_roc_auc = b.RocAuc()(gt_y=y, pred_probas=rf)
# block registration
nn_func_block_wrapper = b.new(NeuralNetwork, dim=dim)
# adding our block in the pipeline
nn = nn_func_block_wrapper(x=X, y=y)
nn_roc_auc = b.RocAuc()(gt_y=y, pred_probas=nn)
stack = b.Stack(['rf', 'nn'], axis=1)(rf=rf, nn=nn)
average = b.Average(axis=1)(X=stack)
roc_auc = b.RocAuc()(gt_y=y, pred_probas=average)
pipeline = b.build(
{'X': X, 'y': y},
{'probas': average, 'rf roc-auc': rf_roc_auc, 'total roc-auc': roc_auc, 'nn roc-auc': nn_roc_auc}
)
GraphvizPainter(figure_dpi=100).from_pipeline(pipeline).render('pipeline.jpeg')
display(Image('pipeline.jpeg'))
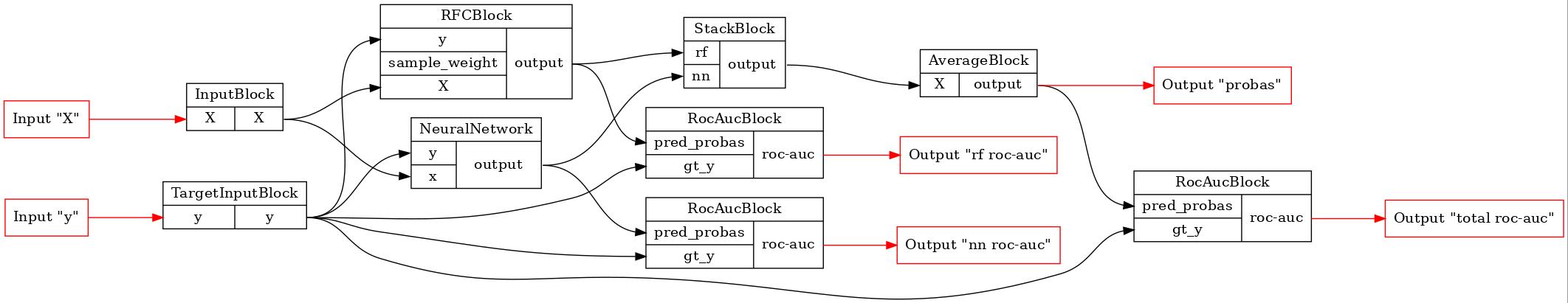
Now let’s fit the pipeline in the parallel mode and see if our ensemble model brought some improvement to the basic models.
[4]:
x, y = make_moons(200, noise=0.5)
train_x, test_x, train_y, test_y = train_test_split(x, y, test_size=0.5)
train_data = {'X': train_x, 'y': train_y}
test_data = {'X': test_x, 'y': test_y}
fit_exec = GreedyParallelExecutor(pipeline, Stage.FIT, outputs=['rf roc-auc', 'nn roc-auc', 'total roc-auc'])
fit_res = fit_exec(train_data).numpy()
for key, val in fit_res.items():
print(f'{key}:', round(val, 3))
rf roc-auc: 1.0
total roc-auc: 0.985
nn roc-auc: 0.923
[5]:
test_exec = GreedyParallelExecutor(pipeline, Stage.TRANSFORM, outputs=['rf roc-auc', 'nn roc-auc', 'total roc-auc'])
test_res = test_exec(test_data).numpy()
for key, val in test_res.items():
print(f'{key}:', round(val, 3))
rf roc-auc: 0.838
total roc-auc: 0.859
nn roc-auc: 0.891