User guide of the models comparison using bosk#
You may want to compare different models in bosk: for example, several bosk deep forests with the different layers number and some models outside of the framework. For this purposes the bosk.comparison subpackage was developed.
This exmaple requires the Pytorch to be installed. Make sure you have the torch package installed or run the following cell:
[ ]:
!pip install torch
Let’s define some models:
[1]:
from bosk.block.zoo.models.classification import RFCBlock
from bosk.comparison.cross_val import CVComparator
from bosk.comparison.base import BaseForeignModel
from bosk.comparison.metric import MetricWrapper
from bosk.data import CPUData, BaseData
from bosk.executor.topological import TopologicalExecutor
from bosk.painter.graphviz import GraphvizPainter
from bosk.painter.topological import TopologicalPainter
from bosk.pipeline.builder.functional import FunctionalPipelineBuilder
from bosk.stages import Stage
from catboost import CatBoostClassifier
from sklearn.datasets import make_moons
from sklearn.metrics import accuracy_score, roc_auc_score
from sklearn.model_selection import KFold
import numpy as np
import torch
from typing import Dict
from torch.utils.data import TensorDataset, DataLoader
from joblib import hash
from IPython.display import Image
[2]:
n_trees = 20
# we will make some similar bosk pipelines
b = FunctionalPipelineBuilder()
X, y = b.Input()(), b.TargetInput()()
rf_1 = b.RFC(n_estimators=n_trees)(X=X, y=y)
et_1 = b.ETC(n_estimators=n_trees)(X=X, y=y)
concat_1 = b.Concat(['X', 'rf_1', 'et_1'])(X=X, rf_1=rf_1, et_1=et_1)
rf_2 = b.RFC(n_estimators=n_trees)(X=concat_1, y=y)
pipeline_1 = b.build({'X': X, 'y': y}, {'output': rf_2})
b = FunctionalPipelineBuilder()
X, y = b.Input()(), b.TargetInput()()
rf_1 = b.RFC(n_estimators=n_trees)(X=X, y=y)
et_1 = b.ETC(n_estimators=n_trees)(X=X, y=y)
concat_1 = b.Concat(['X', 'rf_1', 'et_1'])(X=X, rf_1=rf_1, et_1=et_1)
rf_2 = b.RFC(n_estimators=n_trees)(X=concat_1, y=y)
stack = b.Stack(['rf_2_1', 'rf_2_2'], axis=2)(rf_2_1=rf_2, rf_2_2=rf_2)
av = b.Average(axis=2)(X=stack)
pipeline_2 = b.build({'X': X, 'y': y}, {'output': av})
b = FunctionalPipelineBuilder()
X, y = b.Input()(), b.TargetInput()()
rf_1 = b.RFC(n_estimators=n_trees)(X=X, y=y)
et_1 = b.ETC(n_estimators=n_trees)(X=X, y=y)
concat_1 = b.Concat(['X', 'rf_1', 'et_1'])(X=X, rf_1=rf_1, et_1=et_1)
rf_2 = b.RFC(n_estimators=n_trees)(X=concat_1, y=y)
et_2 = b.ETC(n_estimators=n_trees)(X=concat_1, y=y)
concat_2 = b.Concat(['rf_2', 'et_2', 'X'])(X=X, rf_2=rf_2, et_2=et_2)
et_3 = b.ETC(n_estimators=n_trees)(X=concat_2, y=y)
pipeline_3 = b.build({'X': X, 'y': y}, {'output': et_3})
pipeline_list = [pipeline_1, pipeline_2, pipeline_3]
To use models, written beneath the bosk, you have to write your own adapter class that inherits the BaseForeignModel interface. This interface requires the model to handle bosk data dictionaries and implement the set_random_state method as a comparison experiment should be reproducible.
Let’s define two such models:
[3]:
class CatBoostModel(BaseForeignModel):
def __init__(self) -> None:
super().__init__()
self.grad_boost = CatBoostClassifier(30, verbose=0)
def fit(self, data: Dict[str, BaseData]) -> None:
self.grad_boost.fit(
data['X'].to_cpu().data,
data['y'].to_cpu().data
)
def predict(self, data: Dict[str, BaseData]) -> Dict[str, BaseData]:
return {'output': CPUData(
self.grad_boost.predict_proba(data['X'].to_cpu().data)
)}
def set_random_state(self, random_state: int) -> None:
self.grad_boost.random_state = random_state
class NeuralNetwork(torch.nn.Module):
def __init__(self, dim: int) -> None:
super().__init__()
self.device = torch.device('cuda')
self.nn = torch.nn.Sequential(
torch.nn.Linear(dim, dim * 10),
torch.nn.ReLU(),
torch.nn.Linear(dim * 10, dim),
torch.nn.Tanh(),
torch.nn.Linear(dim, 1),
torch.nn.Sigmoid()
).to(self.device)
self.optimizer = torch.optim.Adam(self.nn.parameters(), 0.0005)
self.batch_size = 64
self.epochs_num = 500
self.loss_fn = torch.nn.CrossEntropyLoss()
def _get_tensor(self, x: np.ndarray) -> torch.Tensor:
return torch.from_numpy(x.astype(np.float32)).to(self.device)
def forward(self, x: torch.Tensor) -> torch.Tensor:
proba = self.nn(x)
return torch.concat((1 - proba, proba), dim=-1)
def transform(self, x: np.ndarray) -> np.ndarray:
with torch.no_grad():
x = self._get_tensor(x)
return self(x).cpu().numpy()
def fit(self, x: np.ndarray, y: np.ndarray) -> 'NeuralNetwork':
x_tens = self._get_tensor(x)
y_tens = self._get_tensor(y).long()
dataset = TensorDataset(x_tens, y_tens)
data_loader = DataLoader(dataset, self.batch_size, True)
self.train()
for _ in range(self.epochs_num):
for x, y in data_loader:
self.optimizer.zero_grad()
pred = self(x)
loss = self.loss_fn(pred, y)
loss.backward()
self.optimizer.step()
return self
class NNModel(BaseForeignModel):
def __init__(self, dim: int) -> None:
super().__init__()
self.nn = NeuralNetwork(dim)
def fit(self, data: Dict[str, BaseData]) -> None:
return self.nn.fit(
data['X'].to_cpu().data,
data['y'].to_cpu().data
)
def predict(self, data: Dict[str, BaseData]) -> Dict[str, BaseData]:
return {'output': CPUData(
self.nn.transform(data['X'].to_cpu().data)
)}
def set_random_state(self, random_state: int) -> None:
torch.manual_seed(random_state)
def init_weights(obj):
if isinstance(obj, torch.nn.Linear):
torch.nn.init.xavier_uniform_(obj.weight)
with torch.no_grad():
self.nn.apply(init_weights)
Also, we need to redefine sklearn metrics a little bit and, just as above, create adapters to handle bosk data dictionaries (in this example we will do it later). Fortunately, in case of simple metrics we can easily use the ready wrapper class MetricWrapper.
[4]:
def accuracy(y_true, y_pred):
return accuracy_score(y_true, np.int_(y_pred[:, 1]))
def roc_auc(y_true, y_pred):
return roc_auc_score(y_true, np.int_(y_pred[:, 1]))
Now we can talk about CVComparator class. It performes various models comparison via cross-validation algorithm. The cross validation scheme is chosen by the user and should have the sklearn BaseCrossValidator interface.
The comparator will try to optimize the bosk pipelines passed in. It means that the comparator will try to find the common part of all pipelines and run this part only once. This will reduce the comparison time. The important detail: common blocks are found using the joblib.hash function and it can produce different md5 hashes for the semantically identical blocks:
[5]:
block_1 = RFCBlock() # 100 trees by default
block_2 = RFCBlock(n_estimators=100) # also 100 trees
# hashes of those blocks will be different
print('- Whether hashes are the same?')
print('-', hash(block_1) == hash(block_2))
- Whether hashes are the same?
- False
Therefore before the comparison for all the blocks across the pipelines the same random seed is set. If for some reason the optimization algorithm ruins your model, you can turn it off by passing False value to the f_optimize_pipelines argument. In this case only set_random_state method will be used for the pipelines before the comparison and the copy.deepcopy during the one.
Let’s draw the changed pipelines to see what’s happening inside of BaseComparator
[6]:
# firstly we will define the CV strategy
# for creating the CVComparator object
random_seed = 42
cv_strat = KFold(n_splits=3, shuffle=True, random_state=random_seed)
# if you don't want to compare foreign models
# or bosk pipelines, just pass None to the
# corresponding argument
comparator = CVComparator(
pipelines=pipeline_list,
foreign_models=[CatBoostModel(), NNModel(dim=2)],
cv_strat=cv_strat,
get_blocks_times=True,
f_optimize_pipelines=True,
random_state=random_seed
)
Found common part of the three pipelines
[7]:
GraphvizPainter(figure_dpi=100).from_pipeline(comparator._common_pipeline).render('pipeline.jpeg')
display(Image('pipeline.jpeg'))
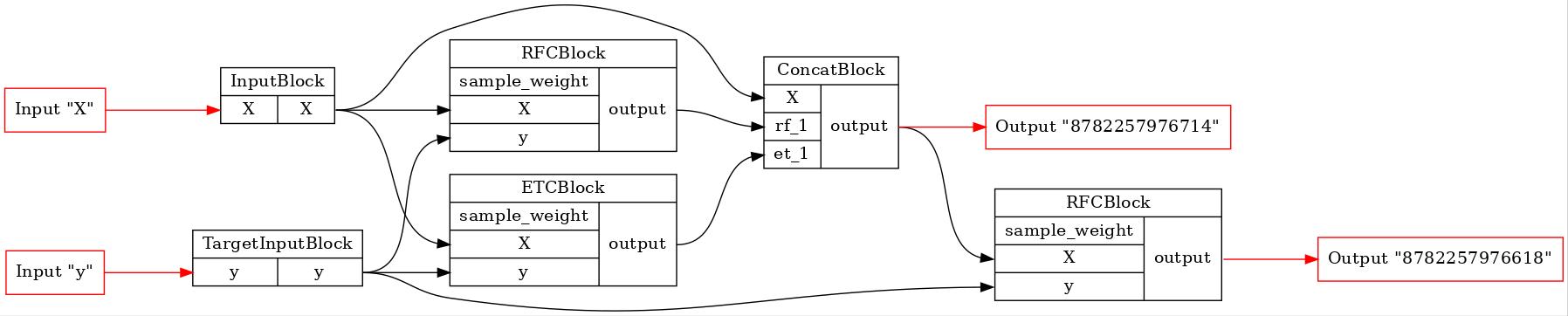
And let’s see the first and the last pipelines on the FIT stage
[8]:
for pip in comparator._optim_pipelines[0::2]:
executor = TopologicalExecutor(pip, Stage.FIT)
TopologicalPainter(figure_dpi=100).from_executor(executor).render('pipeline.jpeg')
display(Image('pipeline.jpeg'))
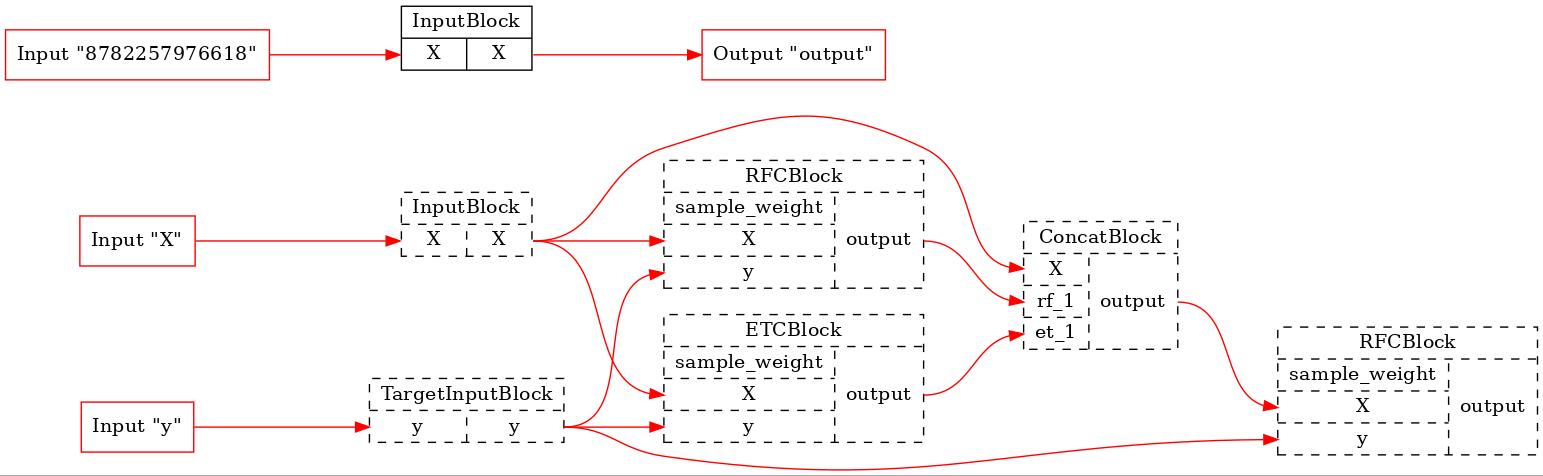
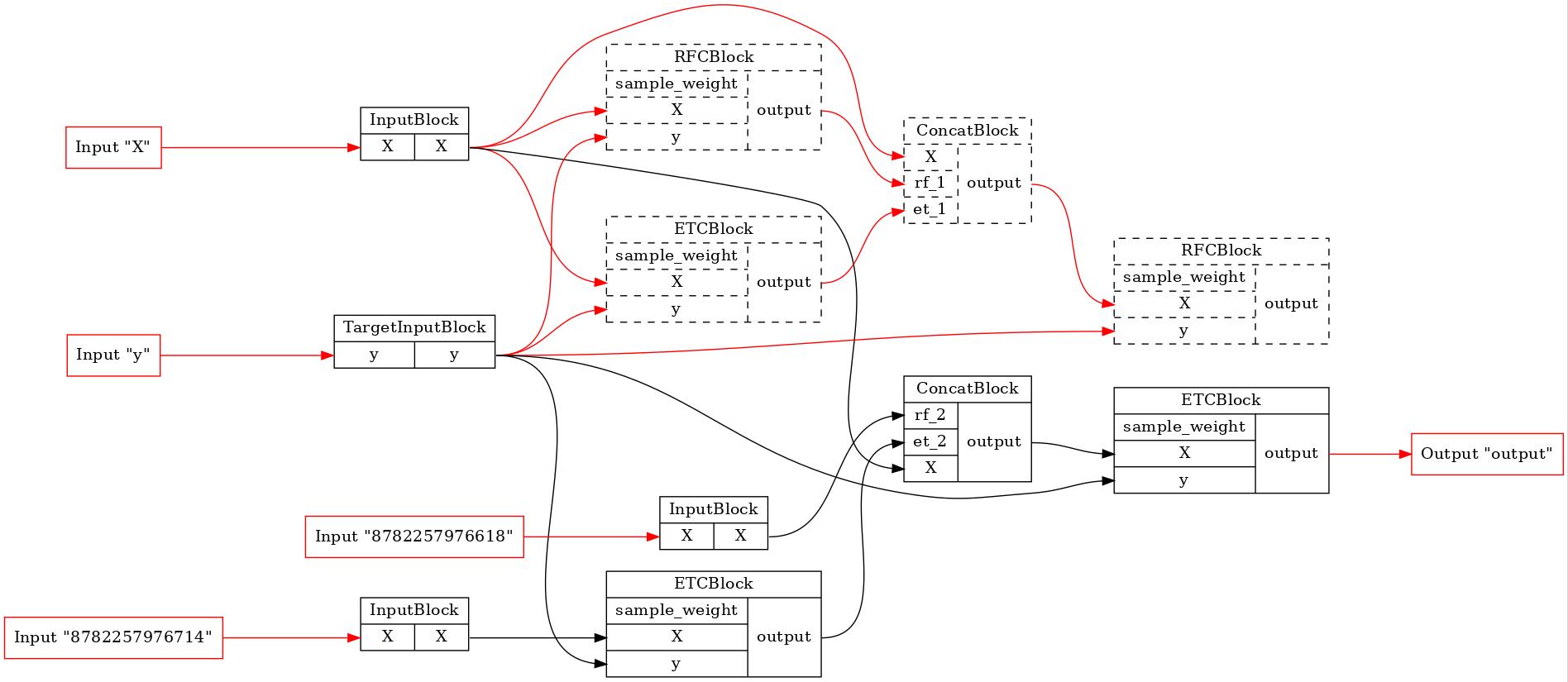
As we can see, none of the original blocks will be executed in the first pipeline. In the last one a lot of blocks were excluded from the training process. So, BaseComparator founds the common part of the pipelines, cuts it out of them and inserts additional input plugs. It allows us run the common part only once and use its results in other pipelines.
Let’s run the comparator and obtain the result table
[9]:
# let's make some data
x, y = make_moons(200, noise=0.5, random_state=random_seed)
data = {'X': CPUData(x), 'y': CPUData(y)}
result = comparator.get_score(data, [
MetricWrapper(accuracy, name='accuracy'),
MetricWrapper(roc_auc, name='roc-auc')
])
result.head()
[9]:
| model name | fold # | train/test | time | blocks time | accuracy | roc-auc | |
|---|---|---|---|---|---|---|---|
| 0 | deep forest 0 | 0 | train | 0.070443 | {InputBlock: 4.89999999953028e-06, TargetInput... | 0.992481 | 0.992857 |
| 1 | deep forest 0 | 0 | test | 0.006083 | {InputBlock: 1.8999999999991246e-06, ETCBlock:... | 0.776119 | 0.772072 |
| 2 | deep forest 1 | 0 | train | 0.070605 | {StackBlock: 6.709999999987559e-05, AverageBlo... | 0.992481 | 0.992857 |
| 3 | deep forest 1 | 0 | test | 0.006156 | {StackBlock: 1.8800000000318562e-05, AverageBl... | 0.776119 | 0.772072 |
| 4 | deep forest 2 | 0 | train | 0.100048 | {TargetInputBlock: 1.4000000003733248e-06, ETC... | 1.000000 | 1.000000 |
We’ve got Pandas Dataframe. Let’s describe all the columns: - model name - is the name of the compared model. In case of the bosk pipeline it will be marked as deep forest i, foreign models are named model i. Models are ordered as they were passed in the initializing list. - fold # - number of the fold starting with 0. - train/test - string binary label if it is a train or test result. - time - CPU time of the fold proceeding. - blocks time - optional column if the
get_blocks_times initialization flag was set to the True. This is the only column containing non-scalar data. The cells contain dictionaries with the pipeline’s blocks as keys and their CPU execution time as values. - metric_name columns - value of the corresponding metric on this fold. If the metric name wasn’t provided, column would be named as metric_i, where i is the position in the initializing list.